Dapeng Wu
NSF Center for Big Learning, University of Florida, Gainesville, FL
Learning While Staying Curious: Entropy-Preserving Supervised Fine-Tuning via Adaptive Self-Distillation for Large Reasoning Models
Feb 02, 2026Abstract:The standard post-training recipe for large reasoning models, supervised fine-tuning followed by reinforcement learning (SFT-then-RL), may limit the benefits of the RL stage: while SFT imitates expert demonstrations, it often causes overconfidence and reduces generation diversity, leaving RL with a narrowed solution space to explore. Adding entropy regularization during SFT is not a cure-all; it tends to flatten token distributions toward uniformity, increasing entropy without improving meaningful exploration capability. In this paper, we propose CurioSFT, an entropy-preserving SFT method designed to enhance exploration capabilities through intrinsic curiosity. It consists of (a) Self-Exploratory Distillation, which distills the model toward a self-generated, temperature-scaled teacher to encourage exploration within its capability; and (b) Entropy-Guided Temperature Selection, which adaptively adjusts distillation strength to mitigate knowledge forgetting by amplifying exploration at reasoning tokens while stabilizing factual tokens. Extensive experiments on mathematical reasoning tasks demonstrate that, in SFT stage, CurioSFT outperforms the vanilla SFT by 2.5 points on in-distribution tasks and 2.9 points on out-of-distribution tasks. We also verify that exploration capabilities preserved during SFT successfully translate into concrete gains in RL stage, yielding an average improvement of 5.0 points.
Sherry: Hardware-Efficient 1.25-Bit Ternary Quantization via Fine-grained Sparsification
Jan 12, 2026Abstract:The deployment of Large Language Models (LLMs) on resource-constrained edge devices is increasingly hindered by prohibitive memory and computational requirements. While ternary quantization offers a compelling solution by reducing weights to {-1, 0, +1}, current implementations suffer from a fundamental misalignment with commodity hardware. Most existing methods must choose between 2-bit aligned packing, which incurs significant bit wastage, or 1.67-bit irregular packing, which degrades inference speed. To resolve this tension, we propose Sherry, a hardware-efficient ternary quantization framework. Sherry introduces a 3:4 fine-grained sparsity that achieves a regularized 1.25-bit width by packing blocks of four weights into five bits, restoring power-of-two alignment. Furthermore, we identify weight trapping issue in sparse ternary training, which leads to representational collapse. To address this, Sherry introduces Arenas, an annealing residual synapse mechanism that maintains representational diversity during training. Empirical evaluations on LLaMA-3.2 across five benchmarks demonstrate that Sherry matches state-of-the-art ternary performance while significantly reducing model size. Notably, on an Intel i7-14700HX CPU, our 1B model achieves zero accuracy loss compared to SOTA baselines while providing 25% bit savings and 10% speed up. The code is available at https://github.com/Tencent/AngelSlim .
Real-world Reinforcement Learning from Suboptimal Interventions
Dec 30, 2025Abstract:Real-world reinforcement learning (RL) offers a promising approach to training precise and dexterous robotic manipulation policies in an online manner, enabling robots to learn from their own experience while gradually reducing human labor. However, prior real-world RL methods often assume that human interventions are optimal across the entire state space, overlooking the fact that even expert operators cannot consistently provide optimal actions in all states or completely avoid mistakes. Indiscriminately mixing intervention data with robot-collected data inherits the sample inefficiency of RL, while purely imitating intervention data can ultimately degrade the final performance achievable by RL. The question of how to leverage potentially suboptimal and noisy human interventions to accelerate learning without being constrained by them thus remains open. To address this challenge, we propose SiLRI, a state-wise Lagrangian reinforcement learning algorithm for real-world robot manipulation tasks. Specifically, we formulate the online manipulation problem as a constrained RL optimization, where the constraint bound at each state is determined by the uncertainty of human interventions. We then introduce a state-wise Lagrange multiplier and solve the problem via a min-max optimization, jointly optimizing the policy and the Lagrange multiplier to reach a saddle point. Built upon a human-as-copilot teleoperation system, our algorithm is evaluated through real-world experiments on diverse manipulation tasks. Experimental results show that SiLRI effectively exploits human suboptimal interventions, reducing the time required to reach a 90% success rate by at least 50% compared with the state-of-the-art RL method HIL-SERL, and achieving a 100% success rate on long-horizon manipulation tasks where other RL methods struggle to succeed. Project website: https://silri-rl.github.io/.
KLIPA: A Knowledge Graph and LLM-Driven QA Framework for IP Analysis
Sep 09, 2025



Abstract:Effectively managing intellectual property is a significant challenge. Traditional methods for patent analysis depend on labor-intensive manual searches and rigid keyword matching. These approaches are often inefficient and struggle to reveal the complex relationships hidden within large patent datasets, hindering strategic decision-making. To overcome these limitations, we introduce KLIPA, a novel framework that leverages a knowledge graph and a large language model (LLM) to significantly advance patent analysis. Our approach integrates three key components: a structured knowledge graph to map explicit relationships between patents, a retrieval-augmented generation(RAG) system to uncover contextual connections, and an intelligent agent that dynamically determines the optimal strategy for resolving user queries. We validated KLIPA on a comprehensive, real-world patent database, where it demonstrated substantial improvements in knowledge extraction, discovery of novel connections, and overall operational efficiency. This combination of technologies enhances retrieval accuracy, reduces reliance on domain experts, and provides a scalable, automated solution for any organization managing intellectual property, including technology corporations and legal firms, allowing them to better navigate the complexities of strategic innovation and competitive intelligence.
KEPLA: A Knowledge-Enhanced Deep Learning Framework for Accurate Protein-Ligand Binding Affinity Prediction
Jun 16, 2025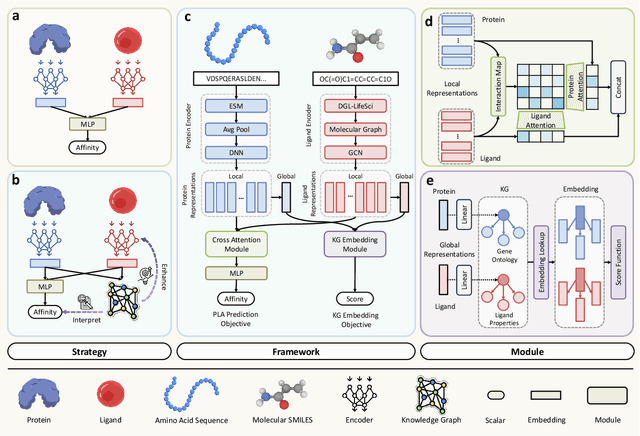
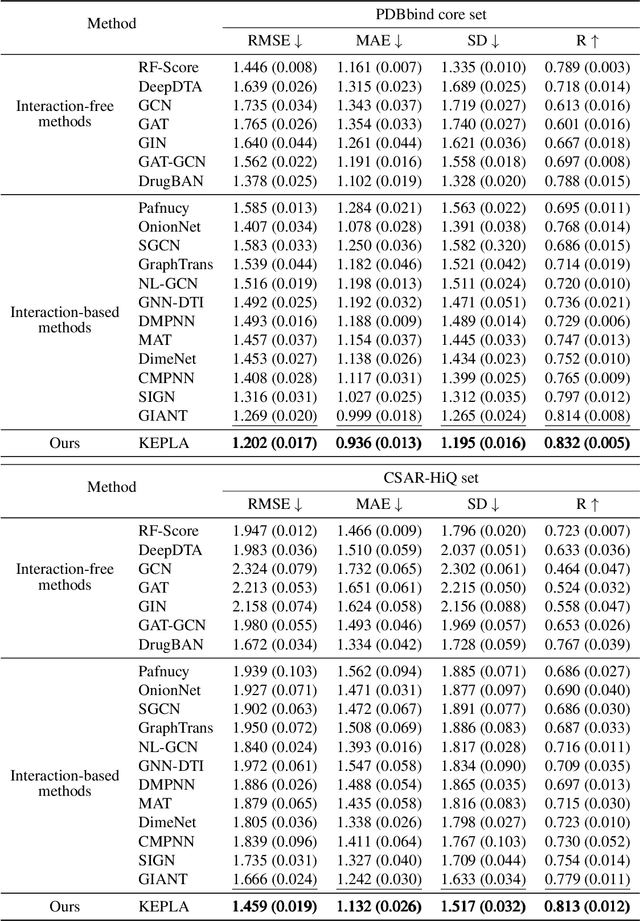
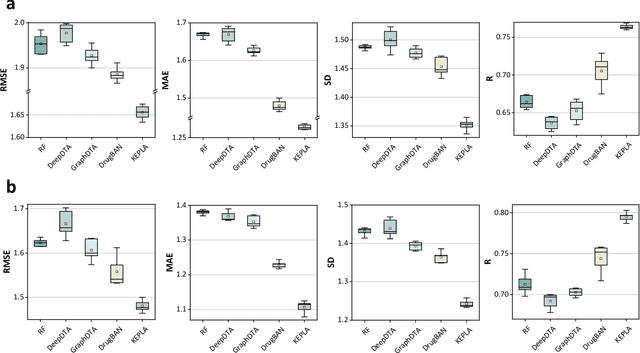
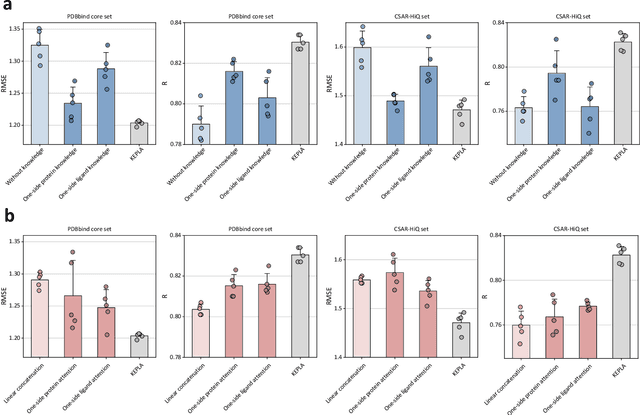
Abstract:Accurate prediction of protein-ligand binding affinity is critical for drug discovery. While recent deep learning approaches have demonstrated promising results, they often rely solely on structural features, overlooking valuable biochemical knowledge associated with binding affinity. To address this limitation, we propose KEPLA, a novel deep learning framework that explicitly integrates prior knowledge from Gene Ontology and ligand properties of proteins and ligands to enhance prediction performance. KEPLA takes protein sequences and ligand molecular graphs as input and optimizes two complementary objectives: (1) aligning global representations with knowledge graph relations to capture domain-specific biochemical insights, and (2) leveraging cross attention between local representations to construct fine-grained joint embeddings for prediction. Experiments on two benchmark datasets across both in-domain and cross-domain scenarios demonstrate that KEPLA consistently outperforms state-of-the-art baselines. Furthermore, interpretability analyses based on knowledge graph relations and cross attention maps provide valuable insights into the underlying predictive mechanisms.
Quaff: Quantized Parameter-Efficient Fine-Tuning under Outlier Spatial Stability Hypothesis
May 20, 2025



Abstract:Large language models (LLMs) have made exciting achievements across various domains, yet their deployment on resource-constrained personal devices remains hindered by the prohibitive computational and memory demands of task-specific fine-tuning. While quantization offers a pathway to efficiency, existing methods struggle to balance performance and overhead, either incurring high computational/memory costs or failing to address activation outliers, a critical bottleneck in quantized fine-tuning. To address these challenges, we propose the Outlier Spatial Stability Hypothesis (OSSH): During fine-tuning, certain activation outlier channels retain stable spatial positions across training iterations. Building on OSSH, we propose Quaff, a Quantized parameter-efficient fine-tuning framework for LLMs, optimizing low-precision activation representations through targeted momentum scaling. Quaff dynamically suppresses outliers exclusively in invariant channels using lightweight operations, eliminating full-precision weight storage and global rescaling while reducing quantization errors. Extensive experiments across ten benchmarks validate OSSH and demonstrate Quaff's efficacy. Specifically, on the GPQA reasoning benchmark, Quaff achieves a 1.73x latency reduction and 30% memory savings over full-precision fine-tuning while improving accuracy by 0.6% on the Phi-3 model, reconciling the triple trade-off between efficiency, performance, and deployability. By enabling consumer-grade GPU fine-tuning (e.g., RTX 2080 Super) without sacrificing model utility, Quaff democratizes personalized LLM deployment. The code is available at https://github.com/Little0o0/Quaff.git.
FedRTS: Federated Robust Pruning via Combinatorial Thompson Sampling
Jan 31, 2025



Abstract:Federated Learning (FL) enables collaborative model training across distributed clients without data sharing, but its high computational and communication demands strain resource-constrained devices. While existing methods use dynamic pruning to improve efficiency by periodically adjusting sparse model topologies while maintaining sparsity, these approaches suffer from issues such as greedy adjustments, unstable topologies, and communication inefficiency, resulting in less robust models and suboptimal performance under data heterogeneity and partial client availability. To address these challenges, we propose Federated Robust pruning via combinatorial Thompson Sampling (FedRTS), a novel framework designed to develop robust sparse models. FedRTS enhances robustness and performance through its Thompson Sampling-based Adjustment (TSAdj) mechanism, which uses probabilistic decisions informed by stable, farsighted information instead of deterministic decisions reliant on unstable and myopic information in previous methods. Extensive experiments demonstrate that FedRTS achieves state-of-the-art performance in computer vision and natural language processing tasks while reducing communication costs, particularly excelling in scenarios with heterogeneous data distributions and partial client participation. Our codes are available at: https://github.com/Little0o0/FedRTS
EPIDetect: Video-based convulsive seizure detection in chronic epilepsy mouse model for anti-epilepsy drug screening
May 31, 2024



Abstract:In the preclinical translational studies, drug candidates with remarkable anti-epileptic efficacy demonstrate long-term suppression of spontaneous recurrent seizures (SRSs), particularly convulsive seizures (CSs), in mouse models of chronic epilepsy. However, the current methods for monitoring CSs have limitations in terms of invasiveness, specific laboratory settings, high cost, and complex operation, which hinder drug screening efforts. In this study, a camera-based system for automated detection of CSs in chronically epileptic mice is first established to screen potential anti-epilepsy drugs.
Spatial-Temporal DAG Convolutional Networks for End-to-End Joint Effective Connectivity Learning and Resting-State fMRI Classification
Dec 16, 2023Abstract:Building comprehensive brain connectomes has proved of fundamental importance in resting-state fMRI (rs-fMRI) analysis. Based on the foundation of brain network, spatial-temporal-based graph convolutional networks have dramatically improved the performance of deep learning methods in rs-fMRI time series classification. However, existing works either pre-define the brain network as the correlation matrix derived from the raw time series or jointly learn the connectome and model parameters without any topology constraint. These methods could suffer from degraded classification performance caused by the deviation from the intrinsic brain connectivity and lack biological interpretability of demonstrating the causal structure (i.e., effective connectivity) among brain regions. Moreover, most existing methods for effective connectivity learning are unaware of the downstream classification task and cannot sufficiently exploit useful rs-fMRI label information. To address these issues in an end-to-end manner, we model the brain network as a directed acyclic graph (DAG) to discover direct causal connections between brain regions and propose Spatial-Temporal DAG Convolutional Network (ST-DAGCN) to jointly infer effective connectivity and classify rs-fMRI time series by learning brain representations based on nonlinear structural equation model. The optimization problem is formulated into a continuous program and solved with score-based learning method via gradient descent. We evaluate ST-DAGCN on two public rs-fMRI databases. Experiments show that ST-DAGCN outperforms existing models by evident margins in rs-fMRI classification and simultaneously learns meaningful edges of effective connectivity that help understand brain activity patterns and pathological mechanisms in brain disease.
scBiGNN: Bilevel Graph Representation Learning for Cell Type Classification from Single-cell RNA Sequencing Data
Dec 16, 2023Abstract:Single-cell RNA sequencing (scRNA-seq) technology provides high-throughput gene expression data to study the cellular heterogeneity and dynamics of complex organisms. Graph neural networks (GNNs) have been widely used for automatic cell type classification, which is a fundamental problem to solve in scRNA-seq analysis. However, existing methods do not sufficiently exploit both gene-gene and cell-cell relationships, and thus the true potential of GNNs is not realized. In this work, we propose a bilevel graph representation learning method, named scBiGNN, to simultaneously mine the relationships at both gene and cell levels for more accurate single-cell classification. Specifically, scBiGNN comprises two GNN modules to identify cell types. A gene-level GNN is established to adaptively learn gene-gene interactions and cell representations via the self-attention mechanism, and a cell-level GNN builds on the cell-cell graph that is constructed from the cell representations generated by the gene-level GNN. To tackle the scalability issue for processing a large number of cells, scBiGNN adopts an Expectation Maximization (EM) framework in which the two modules are alternately trained via the E-step and M-step to learn from each other. Through this interaction, the gene- and cell-level structural information is integrated to gradually enhance the classification performance of both GNN modules. Experiments on benchmark datasets demonstrate that our scBiGNN outperforms a variety of existing methods for cell type classification from scRNA-seq data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge